Universidad Estatal de Bolívar, Facultad de Ciencias Agropecuarias Recursos Naturales y
del Ambiente, Vicerrectorado de Investigación y Vinculación, Guaranda, 020150, Ecuador.
Corresponding author email: fbayas@ueb.edu.ec
Article Publishing History
Received: 25/05/2025
Accepted After Revision: 01/07/2025
Cultivation of strawberry has acquired great importance for consumption, promoting the increase of its production in Ecuador. However, the process of importing plant material from producer countries to improve domestic production has contributed to the dissemination of the fungus Fusarium oxysporum f. sp., fragariae. The objective of this study was to identify the presence of the pathogen by the application of molecular methods to Fusarium strains isolated in the province of Pichincha. Ninety-two diseased strawberry plants and 92 asymptomatic plants were analyzed. Of these samples, 13 fungal isolates with the characteristics of the genus Fusarium were identified. The isolates were analyzed at the molecular level by Polymerase Chain Reaction (PCR) through the amplification of the ITS regions of the rDNA sequences and the EF-1 is a gene, these were sequenced to elucidate the phylogenetic relationships between the isolates. Twelve strains were identified as F. oxysporum f. sp., fragariae. So, the present research contributes to the search for control alternatives to avoid the dissemination of the pathogen in strawberry plantations in Ecuador.
Molecular characterization, Fusarium oxysporum, strawberry
Morejón F. B, León A. T. Molecular characterization of Fusarium oxysporum f. sp. fragariae isolated from strawberry crops Fragaria ananassa Duch in Ecuador . International Journal of Biomedical Research Science. 2025;01(1).
Morejón F. B, León A.T. Molecular characterization of Fusarium oxysporum f. sp. fragariae isolated from strawberry crops Fragaria ananassa Duch in Ecuador. International Journal of Biomedical Research Science. 2025;01(1). Available from: <a href=”https://shorturl.at/4QMIE“>https://shorturl.at/4QMIE</a>
INTRODUCTION
The strawberry (Fragaria ananassa) has become a very important industrial crop worldwide, it can be said that this plant has the most varied and complex management possibilities, which has allowed for unusual development in the productive areas. The strawberry demand in the world is increasing, not only because of its flavor but also because of its health benefits. Markets like Europe and the United States register a high consumption, and most of the worldwide production of this fruit originates there [1]. In Ecuador, intensive strawberry cultivation began in 1983, mainly in the province of Pichincha [2], and in recent years the cultivated area has increased to 1,200 ha in the main strawberry varieties Albion, Monterrey and Diamante in greater proportion in the province of Pichincha with 400 ha, Tungurahua 240 ha and in the provinces of Chimborazo, Cotopaxi, Imbabura and Azuay with 40 ha, being thus the livelihood of the majority of fruit growers in these sectors, which are cultivated between 1300 and 3600 m above sea level with average temperatures of 15 °C. In the country, 12% of the strawberries harvested are exported while the rest of the production is used to meet domestic demand, which means the international market has a large field for exploitation [3].
The specific pathogenicity of Fusarium oxysporum f. sp., Fragariae causes a negative impact on agriculture worldwide as they are causative agents of vascular wilt and basal rot of a large variety of plants, in Ecuador this fungus affects the production of fruit crops such as babaco [4], strawberry, kidney tomato, among others, attributed to the difficult diagnosis of its worldwide dissemination [5].
Currently, the development and use of new molecular biology technologies based on Polymerase Chain Reaction (PCR) can be used for the application of molecular markers [6]. Restriction Fragment Length Polymorphism (RFLP), Random Amplification of Polymorphic DNA (RAPDs), Amplified Fragment Length Polymorphism (AFLPs), among others, are used in the analysis of genetic DNA polymorphism, which in conjunction with the development of bio-informatic software designed for the analysis of sequences, allows to obtain efficient and timely information applicable to a large number of samples, being more specific, objective, sensitive and faster techniques than traditional methods [7]. The main objective of this research is to identify strains of Fusarium oxysporum f. at the molecular level using the ITS region and the ET-1 gene.
MATERIAL AND METHODS
Sampling was carried out in strawberry producing areas located in the province of Pichincha, from which 92 symptomatic and 92 asymptomatic plants were obtained; these plants were in phases of vegetative development and fruiting. The analyses were conducted in Agrocalidad laboratories according to the following procedure:
Seeding of plant material and substrate: Longitudinal cuts of the stem were made and 1-2 cm segments containing areas with and without vascular wilt lesions were removed. After this process, three sets of four segments were seeded in PDA medium, then incubated at 24 °C for 7 days. Isolates which present asexual structures of Fusarium sp., were selected [8].
Preservation of the pathogen: In the Petri dishes of monosporic cultures, 10 mL of sucrose (10%) and 10 mL of peptone (20%) were added with a handle (Digralsky), the mycelium and the poured liquid were homogenized. Paper squares (0.5 cm2) were placed in the solution, letting it rest 8 minutes; then enough paper segments embedded in the dilution were introduced into eppendorf tubes and cooled to 6 °C. After massification, enough biomass was obtained for the extraction of DNA [9, 10].
Molecular analysis (phylogenetic test): For this analysis, DNA was extracted from the mycelium using liquid nitrogen by macerating it in the mortar. The maceration was deposited in eppendorf tubes by adding 1 mL of extraction buffer and 0.5 mL of chloroform, iso-amino alcohol in a 24: 1 ratio. The tubes were introduced into the bain-marie, at 55 °C for 30 minutes while inverting them every 10 minutes. They were let to rest for 5 minutes, then centrifuged at 14000 rpm for 10 minutes. The supernatant was transferred to a new sterile tube, adding 1 mL of isopropanol. It was allowed to freeze at −20 °C for 20 minutes, then centrifuged for 3 minutes at 10000 rpm. When the pellet was formed, rinsings were performed with 70% ethanol once or twice, then the pellet was allowed to dry for 25 minutes and inoculated into TE buffer and RNAse, then left in the environment for 30 minutes. DNA purity was assessed using a specific miniaturized spectrophotometer (nanodrop, Roche LC1536).
Amplification analysis of the gene of interest by PCR: The primers EF1, EF2, ITS1, ITS4 were used in this study (table 1).
Table 1. Nucleotide sequences used for the amplification of ITS (s) and TEF-1α regions
| Region | Initiator | Sequence | Author |
| Internal Transcribed Spacer (ITS) | ITS-1
ITS-4 |
5’-TCCGTAGGTGAACCTGCGG-3’
5’-TCCTCCGCTTATTGATATGC-3’ |
Martin and Rygiewicz (11) |
| Translation Elongation Factor (TEF-1α) | EF-1
EF-2 |
3’-ATGGGTAAGGARGACAAGAC-5’
3’-GGARGTACCAGTSATCATGTT-5’ |
O’Donnell et al. (12) |
For each PCR reaction, a volume of 25 μL was used; with concentrations of 1.5 mM MgCl2, 0.2 mM of each dNTP; 0.5 μM of each primer 1.25 U / 25 μL Taq polymerase (FLEXI), PCR buffer (Green FLEX 1X). The amplification was carried out in a thermocycler according to the following conditions: initial denaturation at 94 °C for 5 minutes, 35 reaction cycles at 94 °C for 30 seconds, annealing at 53 °C for 1minute, initial extension at 72 °C for 1 minute and a final extent at 72 °C for 10 minutes. The PCR product was analyzed by 0.5% agarose gel electrophoresis previously prepared in buffer TBE 1X (Trisborate, EDTA), a molecular weight marker of 100 bp was used and the gel was run for 30 minutes at 100 volts. The gel was then visualized in a UV light transilluminator. PCR products with nonspecific bands were purified by means of the Band-stab PCR process until clear bands were obtained. All amplified were sent to sequence MACROGEN (Seoul, Korea); the results were compared with the GenBank database [13]. Sequences were compared using a BLAST (Basic Local Alignment Search Tool) alignment that allowed gender and species identification (www.ncbi.nlm.nih.gov/BLAST).
RESULTS AND DISCUSSION
After isolation in PDA medium confirmed by microscopic morphology, only 52 samples showed contamination by the fungi: Pestalotia sp., Fusarium sp., Rhizoctonia sp., and Mycosphaerella sp. The presence of spores belonging to the fungus Pestalotia sp., was evidenced in 46 isolates (88.52%); the presence of asexual structures of Fusarium sp., was determined in 13 isolates, representing 25% of the total samples; a minimum percentage (1.52%) with the fungus Rhizoctonia sp., and after analysis of the leaf area of the straw four isolates of Mycosphaerella sp., were identified (7.69%). It should also be noted that there were samples from which more than one isolate was obtained.
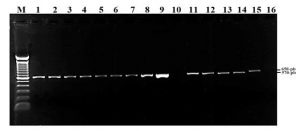
Phylogenetic analysis based on ITS fragments: It was carried out by a PCR with primers ITS1 – ITS4, DNA of the 13 treatments and two possible pathogens was characterized by optical microscopy as Pestalotia sp., and Rhizoctonia sp. A band was evidenced in the gel at a height of 570 base pairs for the 13 treatments, including Pestalotia sp. In the case of Rizhoctonia sp. the band has a slightly higher molecular weight equivalent to 650 bp, Treatment AB8 and negative control (C-) showed no visible band (Figure 1).
Figure 1: Amplified fragment of ITS regions from 13 treatments. Lane M: molecular weight marker (100-bp); lanes 1: AB12, 2: AB13, 3: AB1, 4: AB2, 5: AB3, 6: AB4, 7: AB5, 8: AB6, 9: AB7, 10: AB8, 11: AB9, 12: AB10, 13 : AB11; lane 14: Pestalotia sp; Lane 15: Rhizoctonia sp. Lane 16: negative control.
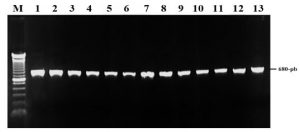
Phylogenetic analysis based on the EF-1α region: The amplification and sequencing of the EF-1α region was performed with the EF1-EF2 primers plus the DNA of the 13 treatments. After PCR amplification, bands of 570 bp were visualized for the 13 treatments and the presence of non-specific bands in each of the isolates was observed, so that the Band-stab PCR technique was performed in all isolates. Amplification of the products extracted with the Band-stab PCR technique enabled the visualization of 680 bp bands in the 13 Treatments (AB1- AB13), with the following DNA concentrations: 80 ng / μl (AB2, AB3, AB4, AB5, and AB8), 100 ng / μL (AB1, AB6, AB7, AB9, AB10, AB11, AB12 and AB13) (Figure 2).
Figure 2: Amplified fragment of EF-1α region of the 13 isolates. Lane M: molecular weight marker (100-bp); lanes 1: AB12, 2: AB13, 3: AB1, 4: AB2, 5: AB3, 6: AB4, 7: AB5, 8: AB6, 9: AB7, 10: AB8, 11: AB9, 12: AB10, 13: AB11.
Other authors have previously studied the ITS and EF-1α regions, demonstrating their effectiveness in solving some generated ambiguities and locating taxonomically new species of the genus Fusarium [14]. Although the use of universal primers allows us to study inter-specific variability and to establish identifications when comparing the sequences obtained with others previously deposited in the GenBank database, it should be emphasized that the best tool to quickly and unequivocally identify at a lower cost the different species of fungi associated with vascular wilt is the use of specific primers (15), but everything will depend on the purity of the isolates and the PCR products, hence several procedures should be performed the best result is reached [16].
Evaluation of the special form of Fusarium oxysporum with specific primers: Specific oligonucleotides designed for Fusarium oxysporum f sp. Fragariae Fofra-1 and Fofra-2 [17], used in the PCR reaction, did not show amplification in the 13 DNA samples. Thus, we must consider that the isolates used in this research did not amplify because there are differences in evolution between species from one locality to another [18, 19].
On the other hand, the ANOVA analysis revealed no significant statistical differences between the treatments in terms of distribution, indicating no variability among the variables. The coefficient of variation was 19.46%, which is considered tolerable and acceptable for field evaluation.
In conclusion, the molecular evaluation identified Fusarium oxysporum f. sp. fragariae as the pathogen responsible for vascular wilt in strawberry plants. During the assessment of pathogenesis, Fusarium oxysporum f. sp. fragariae was detected in all 13 treatments exhibiting vascular wilt symptoms, confirming its presence in Ecuador. This study marks a significant step in recognizing the microbiota associated with strawberry cultivation in Ecuador. It underscores the importance of using molecular techniques for pathogen detection. These tests revealed a high level of diversity among the identified fungal genera and species, providing valuable information for diagnosis and management strategies in strawberry farming.
ACKNOWLEDGMENTS
Thanks to the State University of Bolivar; Faculty of Agricultural Sciences, Natural Resources and Environment, Research Department. Biotechnological Develop and Research Centre, and to the debt exchange program Ecuador – Spain, for the support received in carrying out the present work.
Conflict of Interest Statement: The authors declare that they have no competing interests.
REFERENCES
- Samtani, J. B., Rom, C. R., Friedrich, H., Fennimore, S. A., Finn, C. E., Petran, A., Wallace, R. W., Pritts, M. P., Fernandez, G., Chase, C. A., Kubota, C., & Bergefurd, B. (2019). The Status and Future of the Strawberry Industry in the United States. HortTechnology hortte, 29(1), 11-24. Retrieved May 18, 2024, from https://doi.org/10.21273/HORTTECH04135-18
- Bejarano, W. (1993). New export products. Manual of the strawberry. 1st Ed. PROEXANT, Quito, EC. 1-17.
- Dias Panimboza, D. (2023). Determinación del efecto de agrozoil sobre la incidencia de necrosis radical de fresa Fragaria x ananassa (Duch) variedad monterrey. Tesis de grado, Universidad técnica de Ambato, pp 78. https://repositorio.uta.edu.ec/bitstream/123456789/38128/1/030%20Agronom%C3%ADa%20-%20D%C3%ADas%20Panimboza%20Damaris%20Pamela.pdf
- Jenner BN, Henry PM. (2022). Pathotypes of Fusarium oxysporum sp. fragariae express discrete repertoires of accessory genes and induce distinct host transcriptional responses during root infection. Environ Microbiol. Oct;24(10):4570-4586. doi: 10.1111/1462-2920.16101.
- Tigre León R.A., Erazo, F., Yánez, D., Bayas-Morejon, F. (2020). Phylogenetic analysis of Fusarium oxysporum f strains isolated from strawberry crops Fragaria ananassa Duch in the province of Pichincha (Ecuador). International Journal of Current Pharmaceutical Research, 12(2), 92–95. https://doi.org/10.22159/ijcpr.2020v12i2.37516.
- Henry PM., Kirkpatrick SC, Islas CM, Pastrana AM, Yoshisato J, et al. (2017) The Population of Fusarium oxysporum sp. Fragariae, Cause of Fusarium wilt of Strawberry, in California. Plant Dis 101(4), 550-556. doi: 10.1094/PDIS-07-16-1058-RE.
- Dawidziuk, A., Koczyk, G., Popiel, D., Kaczmarek, J and Buśko, M. (2014). Molecular diagnostics on the toxicogenic potential of Fusarium plant pathogens. J. Appl. Microbiol. 116(6):1607-1620. doi: 10.1111/jam.12488
- Chan, S.D., Jeon, Y.J., Ahn, G.R., Han, J., Han, K., Kim, S.H. (2007). Characterization of Fusarium oxysporum Isolated from Paprika in Korea. Mycobiology 35(2), 91-96. doi: 10.4489/MYCO.2007.35.2.091.
- Henao-Henao, E. D., Hernandez-Medina, C. A., Salazar-González, C., Velasco-Belalcazar, M. L., & Gómez-López, E. D. (2018). Molecular identification of Fusarium isolates associated with passion fruit in five locations from Valle del Cauca, Colombia. Agronomía Mesoamericana, 29(1), 53–61. https://doi.org/10.15517/ma.v29i1.27114.
- Osama, A., Al-Bedak, Rania, M., Sayed, Sedky, H.A. Hassan, (2019). A new low-cost method for long-term preservation of filamentous fungi, Biocatalysis and Agricultural Biotechnology, Volume 22, 101417. https://doi.org/10.1016/j.bcab.2019.101417.
- Martin, Jand Rygiewicz, P.T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol 5(28), 11. https://doi.org/10.1186/1471-2180-5-28.
- O’Donnell, K.,Kistler, H.C., Cigelnik, E., Ploetz, R.C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Natl. Acad 95, 2044–2049. doi: 10.1073/pnas.95.5.2044.
- NCBI BLAST. https://blast.ncbi.nlm.nih.gov/Blast.cgi
- Boutigny, A.L., Gautier, A., Basler, R., Dauthieux, F., Leite, S., Valade, R., Aguayo, J., Ioos, R., Laval, V. (2019). Metabarcoding targeting the EF1 alpha region to assess Fusarium diversity on cereals. PLoS One. Jan 11;14(1):e0207988. doi: 10.1371/journal.pone.0207988.
- Lücking, R., Hawksworth, D.L. (2018). Formal description of sequence-based voucherless Fungi: promises and pitfalls, and how to resolve them. IMA Fungus 9, 143–165. https://doi.org/10.5598/imafungus.2018.09.01.09.
- Mbareche, H., Veillette, M., Bilodeau, G., Duchaine, C. (2020). Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. Peer J. Feb 17;8:e8523. doi: 10.7717/peerj.8523. PMID: 32110484; PMCID: PMC7032056.
- Suga, H., Hirayama, Y., Morishima, M., Suzuki, T., Kageyama, K., Hyakumachi, M. (2013) Development of PCR primers to identify Fusarium oxysporum sp., fragariae. Plant Dis 97, 619-625. doi: 10.1094/PDIS-07-12-0663-RE.
- Jenkins, S., Taylor, A., Jackson, A.C., Armitage, A.D., Bates, H.J., Mead, A., Harrison, R.J., Clarkson, J.P. (2021). Identification and Expression of Secreted In Xylem Pathogenicity Genes in Fusarium oxysporum sp. pisi. Front Microbiol. Apr 9;12:593140. doi: 10.3389/fmicb.2021.593140. PMID: 33897626; PMCID: PMC8062729.
- Peruzzo, A.M., Pioli, R.N., Martínez, L.P., Hernández, F., & Cairo, C.A. (2023). Biological relationships among Fusarium graminearuml. Isolates from diverse hosts and environments of Argentina. Chilean journal of agricultural & animal sciences, 39(2), 217-227. https://dx.doi.org/10.29393/chjaa39-19brac50019.



